
|
Combining the different kinds of current high-throughput data produces new systems-level hypotheses about gene function and regulation, and ultimately functioning of biological organisms. We develop probabilistic modeling, statistical data analysis and machine learning methods to advance this field. We currently have four main focus areas: Systems-level translational medicineTranslational medicine refers to translating molecular-level models and inferences to the patient level. We develop methods for the next step of translational medicine where the goal is to translate systems-level molecular understanding from model organisms to humans. This includes fusing of metabolomics and transcriptomics data, discovery of disease effects, and their mapping between tissues and organisms with probabilistic methods. The initial disease focus is on Type I diabetes. A related project:
Genomics of human endogenous retrovirusesAbout eight per cent of human DNA consists of remains of specific kinds of transposons called human endogenous retroviruses (HERV). Human retroviruses, such as HIV, in general are viruses capable of copying their genetic code to the DNA of humans, and they become endogenous once they have been copied to the germ-line. Human endogenous retroviruses are remains from ancient infections, and it has been suggested that they may have functions in regulating the activity of human genes, and may produce proteins under some conditions. We developed and applied methods for exporing the class structure of HERVs, as well as their association to expression with statistical models, like mixtures of Hidden Markov Models. A related project:
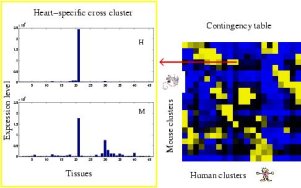
Data fusion for systems biologyA major component of systems biology is integration of information from multiple sources. For example, in cancer it is known that some of the gene expression changes are due to copy number changes in the genome. Both gene expression and copy number changes can be measured, but to find the interesting dependencies between the two, sophisticated integration is required. Another example is gene expression in man and mouse, where we wish to find genes and gene groups with either different or similar activity in the two organisms, in order to study which properties of mice generalize to man. This subfield could be called comparative functional genomics.We have introduced methods for focusing on relevant variation in several data sets, the relevance being determined by auxiliary data sets (for example Gene Ontology classes) or symmetrically by several sets in data fusion. Probabilistic data fusion, mutual dependency modeling, and learning metrics methods (See Statistical machine learning and data mining) provide state of the art tools for this. Related projects:
Information visualization for high-throughput dataThe large and high-dimensional high-throughput data sets are prime application areas for information visualization and other informatics methods. We have so far visualized gene interaction graphs, where the task is to make the huge graphs understandable through visualization. The second application area has been to construct visual interfaces to gene expression databanks. A large community-resource or private gene expression databank consists of numerous data sets submitted by several parties. A key challenge is how to best use the databanks to support further research. Information visualization methods produce an interface to the databank which highlights visually similarities and differences of the data sets.OtherPreliminary joint projects with several other groups.
|
|
This material is presented to ensure timely dissemination of scholarly and technical work. Copyright and all rights therein are retained by authors or by other copyright holders. All persons copying this information are expected to adhere to the terms and constraints invoked by each author's copyright. In most cases, these works may not be reposted without the explicit permission of the copyright holder.
Back to the main page of the research group.You are at: CIS → Research on Bioinformatics
Page maintained by jve at cis.hut.fi, last updated Friday, 24-Sep-2010 10:40:25 EEST